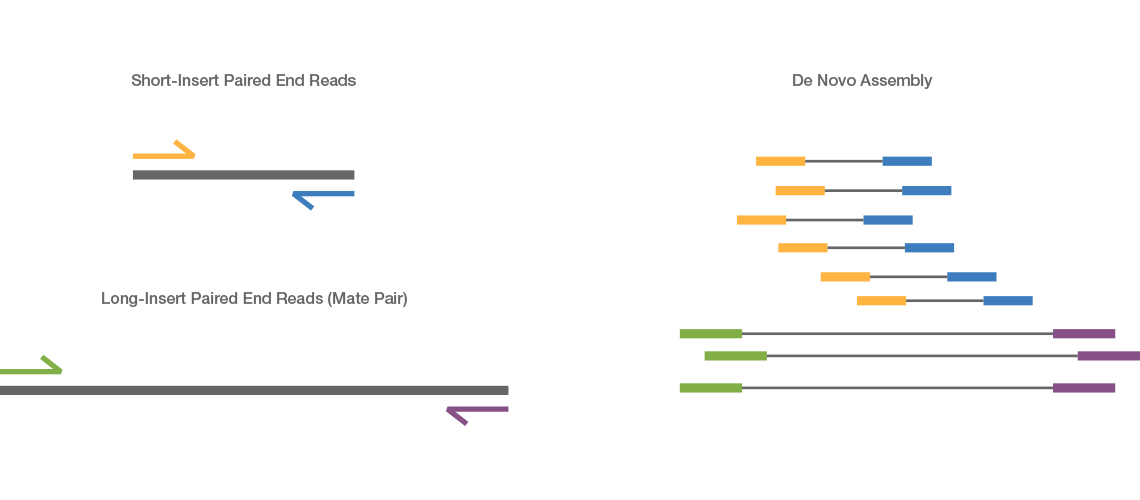
paired end sequencing insert size
Techniques and protocol discussions on sample preparation library generation methods and ideas. 23722180 paired in sequencing 11861090 read1 11861090 read2 0 properly paired 000 23722180 with itself and mate mapped.
Read files from paired-end sequencing need to be paired in Geneious before the pairing information can be used in assembly.
. As we know by now and the insert size limitation becomes an issue when you want paired-end reads with a higher inner distance with fragments longer than 800 bp. The Illumina NexteraXT transposon protocol is a cost effective way to generate paired end libraries. Coverage depth refers to the average number of sequencing reads that align to or cover each base in your sequenced sample.
We developed a new method for long paired-end sequencing of large insert libraries which can efficiently improve the quality of de novo genome assembly and identify. NGS Read Length and Coverage. How to calculate average insert size for paired end reads.
The insert is normally the stretch of sequence between the paired-end adapters so in your case the insert size would be 250 bp 2x75 bp reads 100 bp unsequenced middle piece. In silico empirical estimation of paired-end insert size. This size depends on the library.
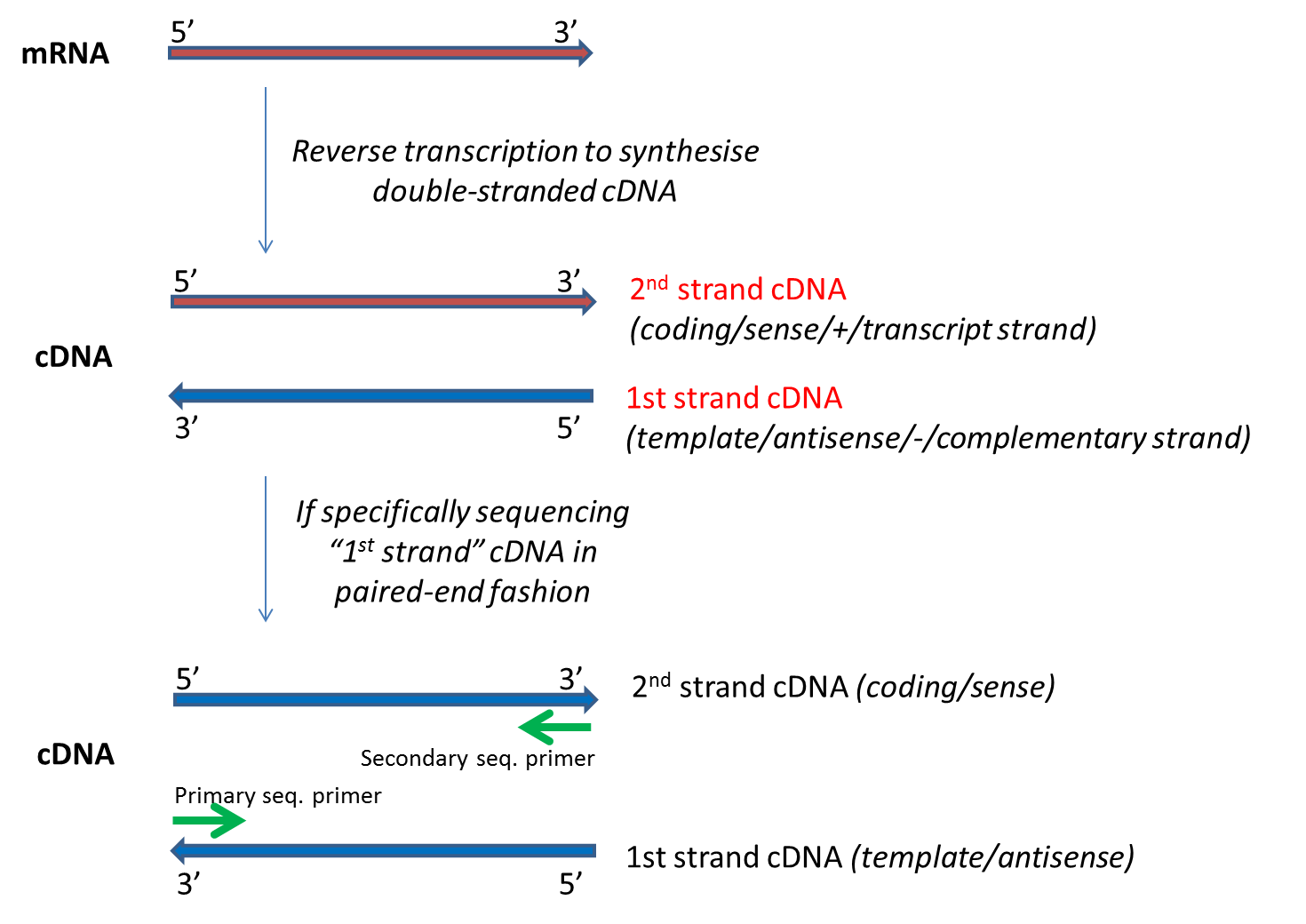
1Create reference transcriptome index for BowTie or BWA. The length of this sequence is known as the insert size not to be. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert.
Before I got my solexa paired end. 2Align paired-end RNA-Seq data to the reference transcripts using minimum insert-length as zero and. Despite the benefits of gaining SS-PE data with paired ends of varying distance the standard Illumina protocol allows only non-strand-specific paired-end sequencing with a single insert.
This can be done using the Set Paired Reads. The reads have a length of typically 50 - 300 bp. - hello there i read this article httpbioinformaticsUconn.
Transpososomes are used to fragment DNA to be sequenced and add. A good choice for read length is closely tied to the insert size of the sequencing library ie how long the individual DNA fragments are that are sequenced.
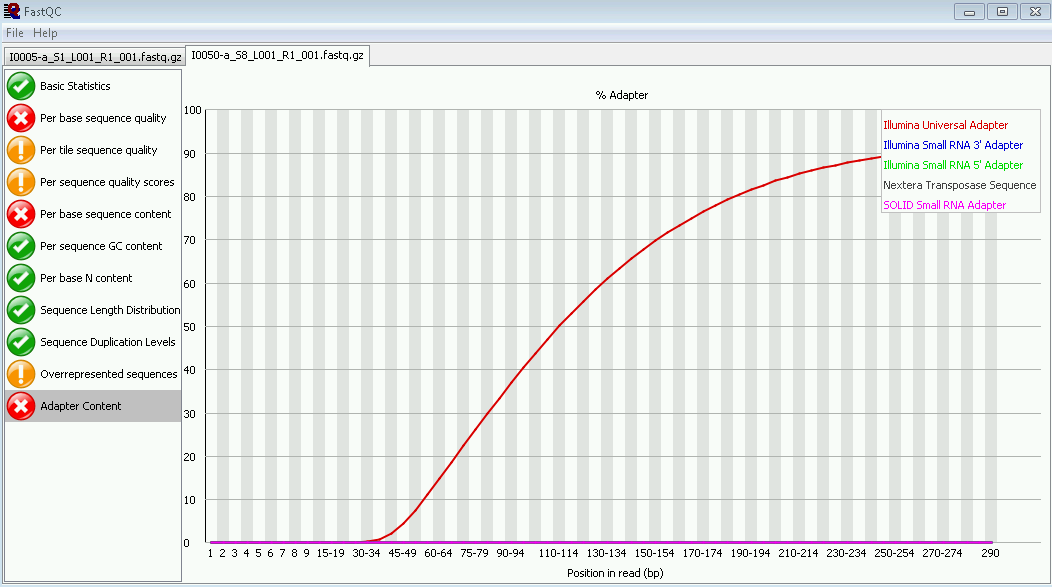
How Short Inserts Affect Sequencing Performance

One End Anchor Reads Due To The Limited Insert Size Orphan Reads Are Download Scientific Diagram

Interpreting Color By Insert Size Integrative Genomics Viewer
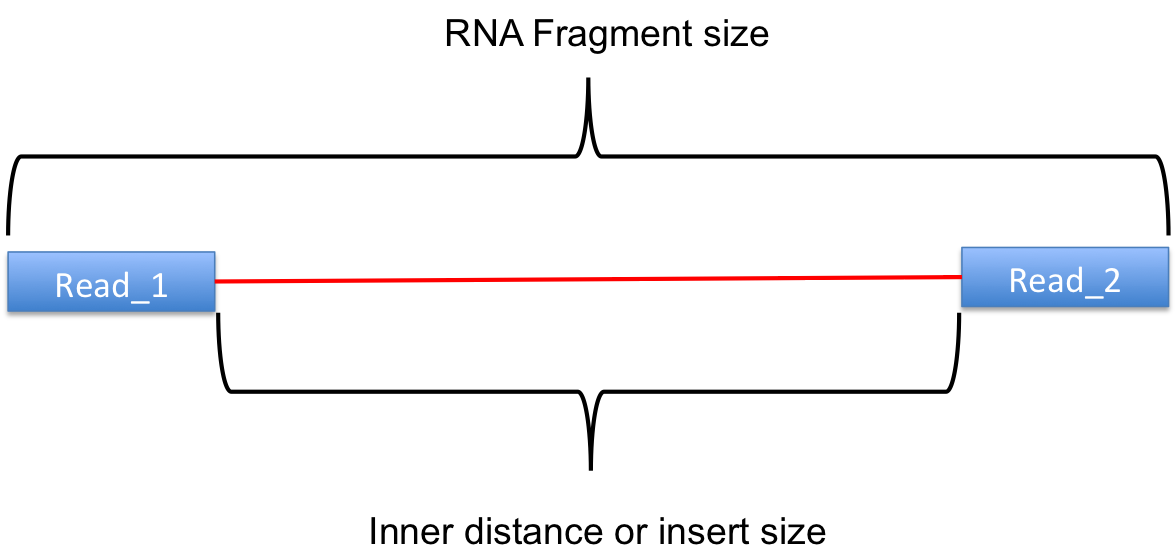
Rseqc An Rna Seq Quality Control Package Rseqc Documentation

Insert Size Metrics Of Atac Seq Data

Detecting Deletion Events A When Mapping Paired End Reads To The Download Scientific Diagram
How Sequencing Works Ngs Analysis
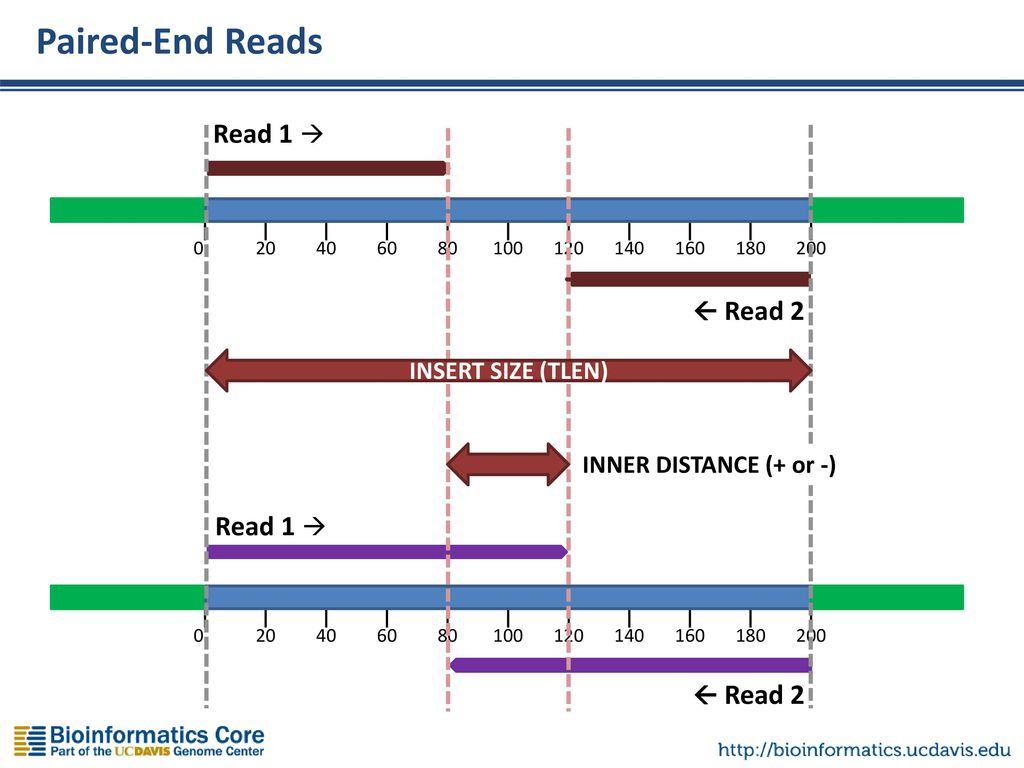
Introduction To Rna Seq Ppt Download
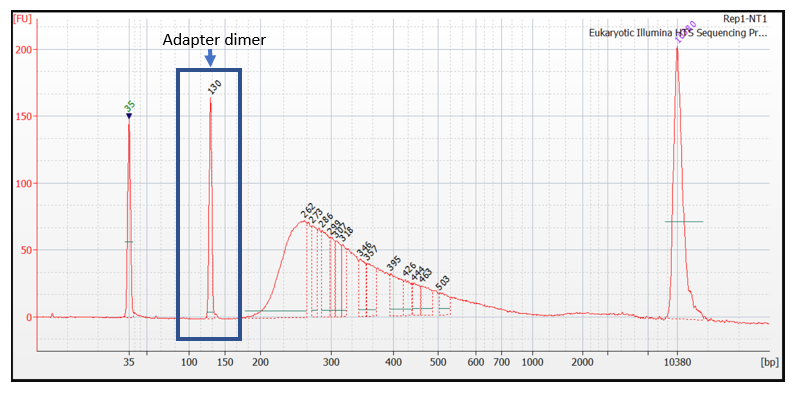
How Short Inserts Affect Sequencing Performance

What Are Paired End Reads The Sequencing Center

Overview Of Pecc Seq A Library Preparation A Modified Pcr Free Download Scientific Diagram

Mp Insert Size Distribution And Library Complexity A Insert Size Download Scientific Diagram
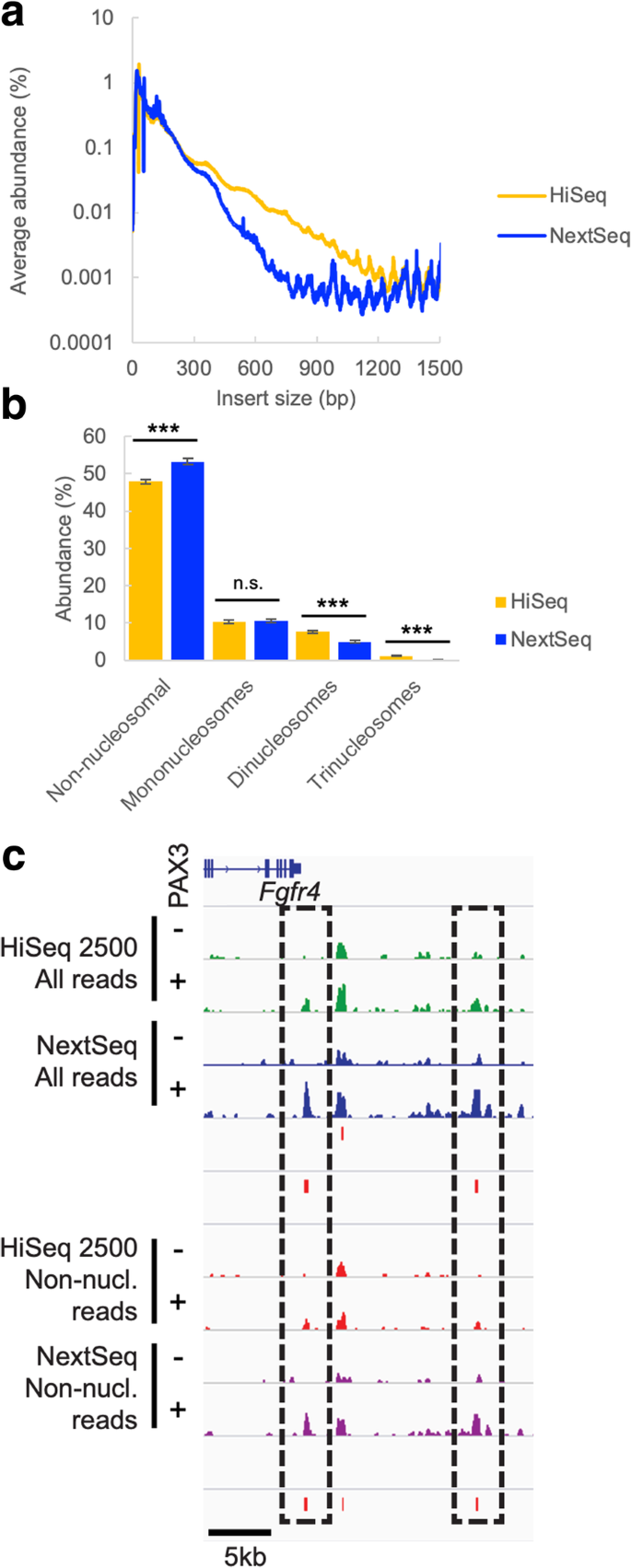
Measuring Sequencer Size Bias Using Recount A Novel Method For Highly Accurate Illumina Sequencing Based Quantification Genome Biology Full Text

Preparation Of Dna Sequencing Libraries For Illumina Systems 6 Key Steps In The Workflow Thermo Fisher Scientific Us
A Genetic Algorithm For Diploid Genome Reconstruction Using Paired End Sequencing Plos One

Illumina Sequencing Illumina Sequencing By Synthesis 1010genome Quality Ngs Bioinformatics Data Analysis Services

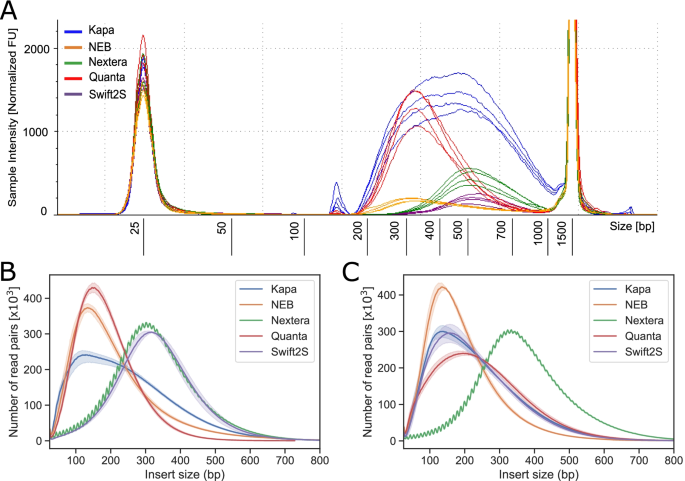
Optimization Of Enzymatic Fragmentation Is Crucial To Maximize Genome Coverage A Comparison Of Library Preparation Methods For Illumina Sequencing Bmc Genomics Full Text